Suppose you have had multiple sensors traverse a section of pipe with the intent of detecting axial cracking. All the major modern tools can detect axial cracking with varying degrees of success. Rather than concern ourselves with which tool does the “best” at detecting the phenomenon, we can treat the signal from each inspection as a piece of evidence supporting or refuting the hypothesis that axial cracking is present. By weighting and combining the opinions of each tool, we can form a more robust assessment that is free of false calls. We see a similar line of reasoning for using ensembles of machine learners, which supply better predictions in aggregate than individually.
I wrote the attached paper called Pipeline Anomaly Assessment: Classifying Gastrointestinal Diseases by their Texture with a Novel Multimodal Classifier as part of my qualifying exam. Excuse the title and the lame introduction. This paper gives a technique for combining multiple sensing modalities and weighting them to best identify a given pathology, like axial cracking.
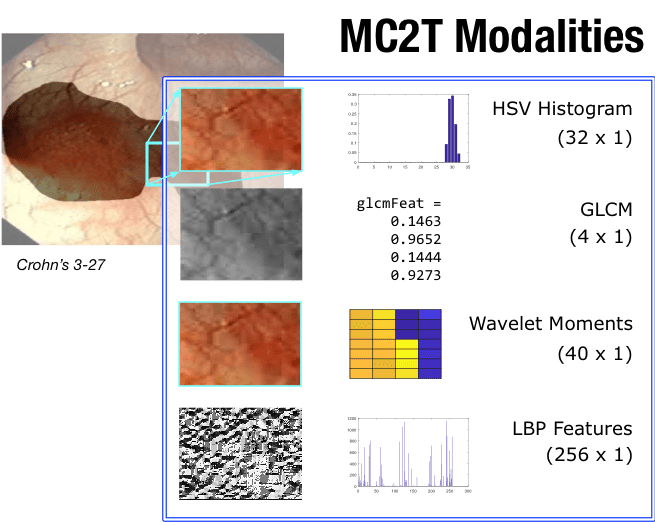
Because I don’t have access to much raw inspection data, I used a database of gastrointestinal diseases called RAGID. While I can’t find a link to the dataset I used, this dataset is very similar. Since these are all video images, I simulated using different types of pipeline sensors by using various image descriptors. With these modifications, my parallel research question became: Can I discriminate various colon diseases better with appropriately weighted sensor modalities than with a single vector that combines the modalities linearly?
In a nutshell, the paper learns how to weight each modality in the context of the pathology I am trying to detect. I get one set of weights per target pathology. Then, I compared the learner’s ability to classify various targets with a simple support vector machine (SVM), where I combined all the modalities into a single stacked vector.
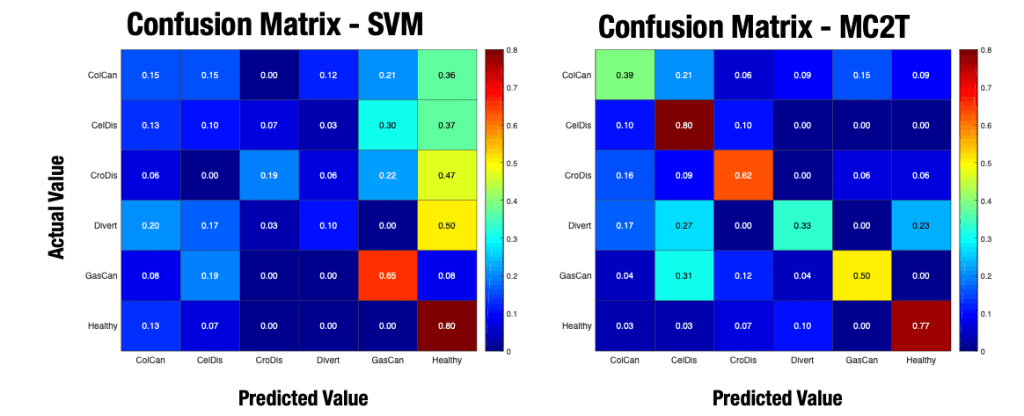
The resulting confusion matrix is the image at the top of this post. The matrix on the left is the SVM with the stacked set of descriptors (sensor modalities). On the right is my custom learner, which learns how to weight each modality to best identify the five target pathologies and healthy tissue. As you can see, the stacked set of descriptors in the SVM didn’t do much except increase the complexity. In all cases, the SVM could not distinguish diseased from healthy tissue. Compare this result with the confusion matrix of my custom machine learner on the right. You can see from the warm colors on the diagonal of this matrix that my custom learner is very good at identifying each pathology using its learned set of modality weights.
Are you able to see the connection to pipelines? Proper signal registration must be addressed before this technique is applicable. I have done a lot of work on registration, so maybe this idea is more relevant than it might first appear?
After digging out this paper, I think I might clean it up and try to get it published. What do you think?
Leave a comment